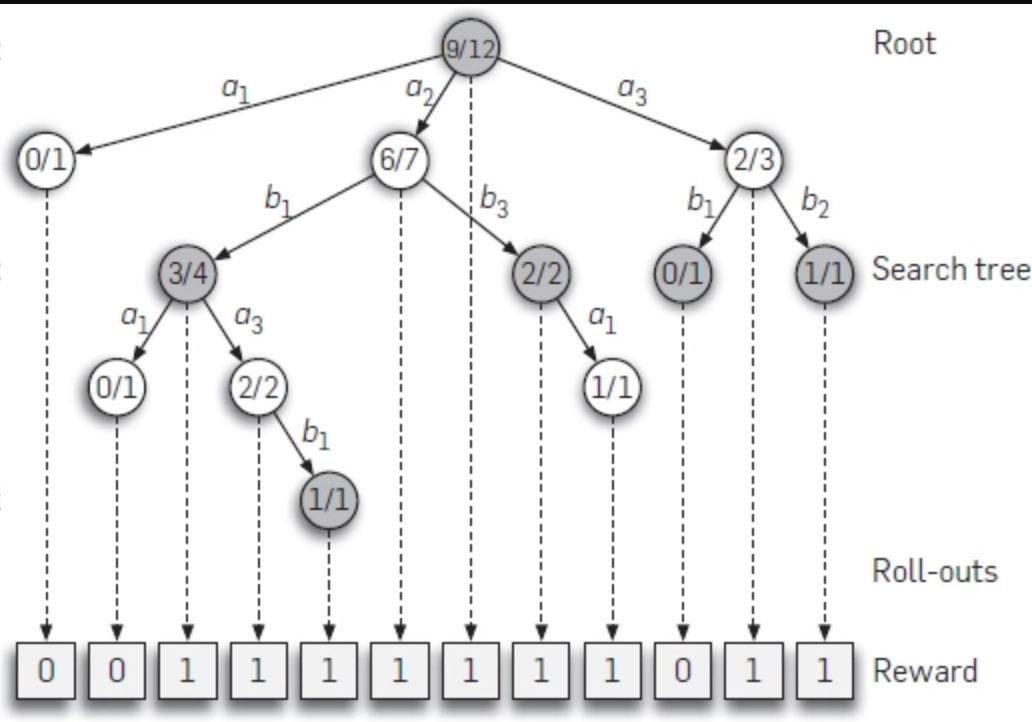
GitHub - petosa/simple-alpha-zero: Clean, tested, & modular
Por um escritor misterioso
Descrição
Clean, tested, & modular AlphaZero implementation with multiplayer support. - GitHub - petosa/simple-alpha-zero: Clean, tested, & modular AlphaZero implementation with multiplayer support.
GitHub - MannLabs/alphapept: A modular, python-based framework for mass spectrometry. Powered by nbdev.

Genome-wide survey of D/E repeats in human proteins uncovers their instability and aids in identifying their role in the chromatin regulator ATAD2 - ScienceDirect
multiplayer · GitHub Topics · GitHub

Proteome‐scale mapping of binding sites in the unstructured regions of the human proteome

Protein Design: From the Aspect of Water Solubility and Stability

Approaches to Heterogeneity in Native Mass Spectrometry

Comprehensive Structure and Functional Adaptations of the Yeast Nuclear Pore Complex
GitHub - dctian/SimpleAlphaZero: A simplified implementation of DeepMind's AlphaZero (with Monte Carlo Tree Search, etc), which can be used to train agents to play simple board games, such as Connect4, Tic-Tac-Toe, and

Heterogeneous non-canonical nucleosomes predominate in yeast cells in situ

Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules
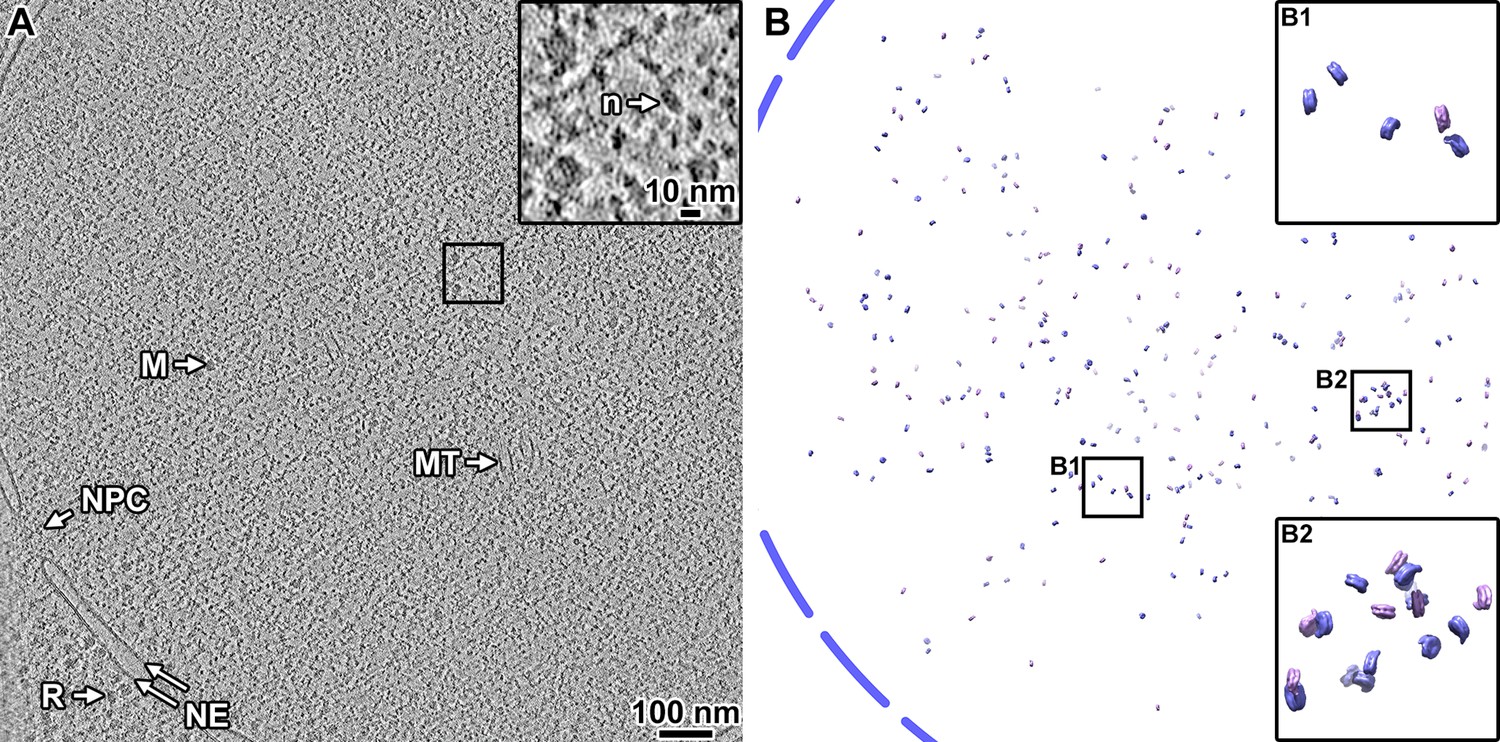
Heterogeneous non-canonical nucleosomes predominate in yeast cells in situ

A practical guide for analysis of histone post-translational modifications by mass spectrometry: Best practices and pitfalls - ScienceDirect

The IbeA protein from adherent invasive Escherichia coli is a flavoprotein sharing structural homology with FAD‐dependent oxidoreductases - Paris - The FEBS Journal - Wiley Online Library
de
por adulto (o preço varia de acordo com o tamanho do grupo)